2013-06-07
Mike Tayeb is the manager of technical support at DNA Genotek.
Several months ago our Vice President of Research and Development, Rafal Iwasiow, wrote a blog discussing the performance of DNA extracted from Oragene/saliva samples in next-generation sequencing applications. In that post, he mentioned validations that demonstrate the suitability of saliva for sequencing that targets particular regions of the genome, such as the whole exome or the major histocompatibility complex (MHC). Also discussed was a study conducted by Dr. Cory McLean of 23andMe regarding the utility of Oragene/saliva for whole genome sequencing (WGS).
As you may already know, the majority of DNA in saliva comes from white blood cells however, human saliva also contains bacteria. When extracting DNA from saliva, some of that bacterial DNA is recovered along with the human DNA. When compared to other oral sampling methods, such as buccal swabs or mouthwash, saliva collected with Oragene contains a substantially lower amount of bacterial DNA with the mean bacterial DNA content being approximately 11%. What I’d like to focus on here is a question that many researchers have regarding the use of saliva for WGS: What is the impact of bacterial DNA in saliva when sequencing the entire genome?
Recently, we conducted a preliminary study to help answer this question. We collected saliva samples using Oragene from a large group of volunteers and extracted the DNA using prepIT•L2P, an ethanol precipitation-based procedure. Three of the individuals also provided blood samples to serve as controls, and DNA was isolated from these using a Qiagen QIAamp DNA Blood Mini Kit. All the DNA samples were quantified using Picogreen and the bacterial DNA content determined using qPCR. From the large set of saliva samples collected, we selected 21 samples that spanned a wide range of bacterial DNA content, from approx 5% to 40%. (It is important to note that samples with bacterial content as high as 40% are rare, however we wanted to include some of these “worst case scenario” samples in this study. Please see this white paper for a more complete discussion about bacterial DNA in saliva samples.)
Sequencing libraries were prepared using Illumina TruSeq DNA kits and barcoded paired-end adapters. The libraries were sequenced on the Illumina HiSeq2000, multiplexed to 8 samples per lane on the flow cell. Alignment of the generated sequences with the reference was performed using Illumina’s CASAVA software.
Not surprisingly we found a direct correlation (R2=0.8) between the amount of bacterial DNA in the samples and the proportion of reads that did not map to the human reference genome (see the chart below). For the blood samples, which were confirmed via qPCR to contain virtually no bacterial DNA, an average of 4% of reads did not align to the human reference and this value was used as a background correction for all samples. For the Oragene/saliva samples, 5.3% of the total reads, on average, after background correction, were unmapped. In the “worst case scenario” sample (bacterial content of 40.3%), 9.9% of the reads did not map to the reference. Interestingly, for all samples, the amount of unmapped reads is lower than the % bacterial DNA estimated by qPCR.
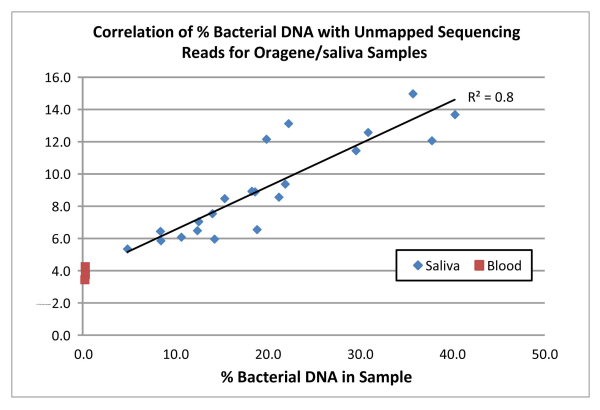
These results indicate that although there is some reduction in the proportion of reads that map to the human reference, that decrease is small, even in samples containing an unusually high amount of bacterial DNA.
We are currently conducting further whole-genome sequencing validation studies of Oragene/saliva samples including a more thorough investigation of the relative performance between blood- and Oragene/saliva-derived DNA using paired samples from the same donor and the effect on variant calling. We expect to have results later this year. If you have any unanswered questions about Oragene/saliva samples and next-generation sequencing (either whole genome or targeted) please email us at info@dnagenotek.com


