2020-05-07
2020 so far has been an interesting year for me. I started the year by attending one of the biggest precision medicine conferences in the world, Precision Medicine World Conference (PMWC). The conference covered topics including immune-oncology, biomarker diagnostics, artificial intelligence, pharmacogenomics, and data science. As a product manager supporting our microbiome lab services and a keen follower of microbiome research, I was particularly interested in the trending topic of the utility of the microbiome.
The set up at PMWC is a bit different from a traditional conference, rather than having concurrent talks hosted in separate rooms, all the talks are hosted on open stages throughout the exhibit hall. It sounds strange but it creates a wonderful atmosphere that encourages networking and makes it easy to bounce from track to track. Throughout the week of the conference, I noticed the talks on the microbiome were packed, which also encouraged others walking by to stop and listen. It was obvious that people were seeking information about microbiome utility and the information you can get from microbiome samples.
Dr. Dan Knights, CEO of CoreBiome (DNA Genotek’s sister company) and Associate Professor at the University of Minnesota, presented ways he and the team at CoreBiome make shotgun metagenomics accessible for large population-scale microbiome studies. As always, Dr. Knights drew a huge crowd.
For those who weren’t lucky enough to hear his talk live (and those who want to hear it again), I am sharing the highlights from Dr. Knights’ presentation, focusing on those I found most interesting and relevant to the microbiome field.
Shotgun metagenomics accessibility for population-scale microbiome studies
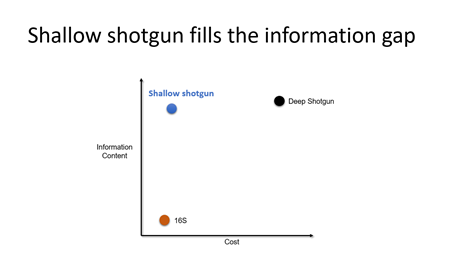
Dr. Knights shared his experiences by applying techniques from machine learning to microbiome sequencing data. In the past, he typically worked with either low resolution 16S rRNA sequencing data that provided genus-level profiles for lots of samples or very high resolution deep shotgun sequencing data which is often too expensive to use in large studies. He wondered if there was a middle ground between these two commonly used sequencing techniques. Dr. Knights and his team at the University of Minnesota developed a technique that they called shallow shotgun sequencing that bridges the gap between 16S amplicon sequencing and deep shotgun at a price point that makes it easier to use this sequencing method in large population-scale studies. This shallow shotgun sequencing technique was one of the discoveries that formed the basis for the company CoreBiome.
“This was kind of revolutionary for us in our own studies because it allowed us to stop doing this two stage system where we would do the 16S first on the whole study and then zoom in with shotgun on a subset of samples. Now we just do a mid-depth or shallow shotgun sequencing study on all the samples.”
Shallow shotgun sequencing: the better alternative?
Dr. Knights described shallow shotgun sequencing as a bridge for the microbiome information gap. In a validation study, he showed data that demonstrated that deep shotgun data and shallow shotgun data can capture similar signals in beta diversity and alpha diversity.
“We can also recover species level biomarkers using shallow sequencing and the significant species between normal and type 2 diabetes using deep and shallow shotgun sequencing.”
Dr. Knights explained that shallow shotgun sequencing provided good profiles for the known genes and the known strains and species. This is thanks to the computational approaches they developed to extract maximum information from the data. They also developed a new alignment algorithm that helps ensure you are calling the best match from the strain database.
The major benefit of shallow shotgun is that it allows researchers working with large cohorts the option of getting data that is higher resolution than amplicon sequencing.
“[It is important to note] that more data is always better. I would never advocate for less data. And if you can afford deep sequencing on your study, you should do it. But the point is, it is usually more important to have more subjects/samples than it is to go deeper on a small number of samples.”
Application example: How to apply shallow shotgun sequencing to your microbiome study

For many of the members of the audience, and possibly even you, there was one obvious question …how can we apply this?
Dr. Knights presented an example from a study led by Dr. Abby Johnson, from the University of Minnesota. The study focused on how daily sampling revealed personalized diet-microbiome associations in humans.
“Diet is the main driver of human gut microbiome variation. Agree or disagree? This is not entirely known or settled in the field, and you can find plenty of evidence for each of these conclusions. We wanted to get to the bottom of this using this new [shallow shotgun sequencing] technology that we had.”
Dr. Abby Johnson and her co-authors had 34 participants collect daily stool samples for 17 days and performed shallow shotgun sequencing on all of these samples. When they analyzed the data they saw the species level profiles in each person changing over time. According to Dr. Knights, he hadn’t seen this level of variation within a person at this level of granularity. You can see the variation in the daily microbiome in the top plot and the variation in the donors’ diet in the bottom plot.
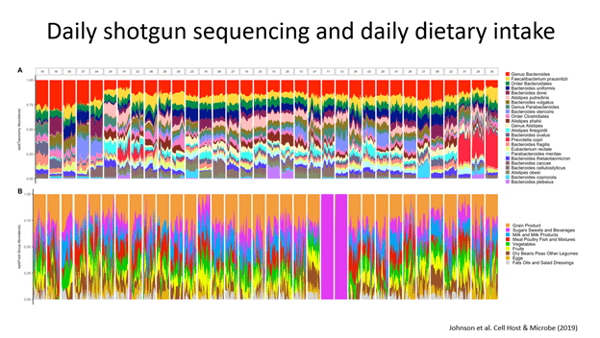
After looking at this data, you might ask – how does the variation of diet compare to the variation of the microbiome? According to Dr. Dan Knights, the diet is more variable than the microbiome.
“And this is looking high level at the diet. What is interesting is that this diet data almost looks like what you get when you do multiomics, but it is food-omics.”
Does diet variation drive microbiome variation?
With this kind of data, you can do all sorts of new things. Dr. Johnson found that microbiome stability (your own microbiome variation from day to day or lack thereof) is very strongly correlated with the diversity of foods you tend to eat on a given day. So the question was, does diet variation drive microbiome variation?
Dr. Knights and his team realized that they had a very tricky problem on their hands because the diet data was so variable it was not clear how they were going to deal with all this variation. Typically researchers break the diet down into nutrient components to reduce the variability they have to work with.
Pairing the average microbiome and nutrient intake
They took the average microbiome of all the participants throughout the study and created a ‘microbiome cloud’, then took the average nutrient intake and created a ‘nutrient cloud’. Using an analysis called Procrustes Analysis they found that their results were a bit of a ‘hairball’ – they did not really correspond.
“Conventionally, the approach has been to break those foods down into nutrients which would be great if we knew everything that was in the foods. But of course, we are only tracking the 100 or so nutrients that we know the human body cares about. We are not tracking what is in the foods that microbes care about.”
After seeing this ‘hairball’ of results, they decided to use food intake instead of nutrients to compare to the microbiome.
Pairing the average microbiome and food intake
To make this analysis possible, Dr. Johnson developed a Food Tree to organize similar foods together and allow Dr. Knights’ team to use analysis tools that leverage phylogenic trees. Using the Procrustes Analysis, they found that the average food intake clouds had a strong correlation to the microbiome clouds and that food intake explained about 19.4% of microbiome variation between people.
They went a step further and leveraged the dense longitudinal shallow shotgun data to discover that response to food was personal. For example, in one study subject cakes and cookies were associated with the growth of Roseburia inulinvorans, which consumes inulin, a common food additive, and in another, the signal would go the other way. This would not have been possible with lower resolution 16S sequencing.
The main point Dr. Dan Knights wanted to leave with the audience that day was that scalable shotgun sequencing can enable new types of studies by providing high resolution and dense longitudinal data.
Like I mentioned earlier, microbiome utility was a hot topic at the PMWC 2020 conference, and Dr. Dan Knights’s talk was, in my opinion, spot on that theme. If you are interested in learning more from his talk you can watch the video above.
If you are interested to learn more about shallow shotgun sequencing, send us an email at info@dnagenotek.com or click here for more information.
Related Blogs
Industry leaders Diversigen and CoreBiome come together to support microbiome discovery
Standardization in microbiome measurements: considerations from sampling to analysis