2020-07-14
The genomic direct-to-consumer, pharmacogenomics and personalized medicine markets are dramatically increasing the number of genomic samples being collected and analyzed. Now with non-invasive collection methods, such as Oragene·Dx and ORAcollect, improved patient access is driving those numbers up even further, placing new challenges on genomic laboratories to process more samples, more efficiently.
In a previous blog we wrote called saliva sample to sequencer in under 8 hours, we explained how our collaboration with Twist Bioscience achieved a single day workflow enabling laboratories to scale up their sample throughput to meet the growing market demands. The collaboration highlighted the time savings from using prepIT·Q2A and Twist’s FAST HYB kit and targeted enrichment technology.
(Read more about DNA Genotek’s collaboration project with Twist)
In this blog, we sat down with Emily Leproust, CEO and co-founder of Twist Bioscience, to chat about the advantages of targeted exome sequencing in a market seeking revolutionary scalable sequencing solutions that deliver fast, inexpensive and accurate genomic data. The interview began by asking Emily for her take on the shift away from microarrays and the increased momentum toward targeted exome sequencing.
Next generation targeted sequencing vs microarrays
With technology advancements and declining costs, the popularity of next-generation sequencing (NGS) has steadily increased and is now challenging microarrays as the tool of choice for genomic analysis. It would seem that the once thought of ‘out of reach’ technology is now readily available for all, making the decision point whether to use one technology over the other a new consideration that must be weighed. From Twist’s perspective, there is a clear advantage to leveraging the targeted sequencing technology.
What benefits does targeted sequencing provide over other downstream analysis methods such as microarrays?
“The quality of sequencing data and the ability to detect a large number of variant types provides a strong advantage over microarrays (i.e. inversions, fusions, and structural variants). Furthermore, hybrid selection capture can detect novel variants and unlocks the speed and flexibility of design change, future-proofing your assay.” – Emily Leproust
Targeted exome sequencing for high throughput labs
Targeted NGS enables rapid identification of common and rare genetic variants. Targeted sequencing can allow for the detection of variants contributing to therapeutic drug response or adverse effects, and is essential for implementing precision medicine.[1] Emily commented that we’re starting to see the positive impacts of targeted sequencing across many healthcare applications.
What healthcare applications can benefit from targeted sequencing?
“All applications benefit dramatically from targeted sequencing. Somatic mutant detection, rare disease investigation, personalized health care for actionable biomarkers, drug monitoring, pathogen detection and early detection of cancer screening are major areas where Twist and targeted sequencing improve health care.” – Emily Leproust
According to a paper published by Polla et al., while exome sequencing has become more common, including for diagnostic purposes, analysis of mutations in disease groups in which the associated genes are largely known has been successfully carried out with a limited number of genes using the targeted panel sequencing approach.[2]
This approach, also known as targeted exome sequencing, uses specific gene panels, focusing only on known loci, allowing increased quality, fidelity, performance and speed. With this approach it is also possible to sequence samples in larger quantities, further decreasing cost due to increased efficiency.[2]
How has the targeted exome sequencing market changed over the past several years?
“[In recent years] technological advances and price pressure of target enrichment has made the entry into this space more appealing than ever. Clinicians and scientists can now plan the experiments to answer questions on a broader scale driving targeted exome sequencing to a larger audience. ” – Emily Leproust
Sample to sequencer in 8 hours: no compromise on quality results
At DNA Genotek and Twist Bioscience, we believe cost effectiveness shouldn’t come at the expense of quality but rather should be the icing on the cake. Through our collaboration, a streamlined 8 hour workflow was presented.
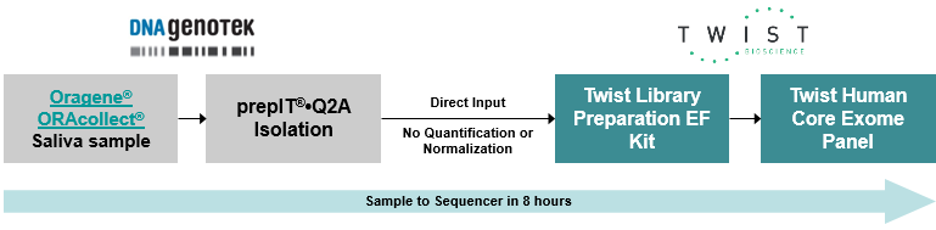
DNA Genotek’s ORAcollect Dx and Oragene Dx collection kits ensured a quality DNA sample entered the workflow, prepIT.Q2A enabled a direct to targeted NGS approach leveraging Twist’s target enrichment solutions. Twist’s FAST HYB library preparation kit offers a robust and tunable library construction method. Along with the Twist Universal Adaptor System, a maximum yield of libraries is generated, forming the foundation for high-throughput sequencing. Emily highlighted the importance of ‘quality in’ to get ‘quality out’.
How important is the workflow before exome sequencing?
“Having a rapid, easy to use, robust, and automatable extraction method for saliva and beyond will provide the needed upstream workflow for targeted sequencing. Coupled with a fast enrichment system, prepIT·Q2A enables a sample to be sequenced in one day with reliable quality.” – Emily Leproust
Together with a more efficient initial workflow using prepIT·Q2A and Twist’s FAST HYB kit, targeted exome sequencing provides a robust and rapid approach to sequencing workflows with high sample throughput, and provides the highest quality data for laboratories sequencing hundreds and even thousands of samples daily.
We believe NGS is here for the long haul and are excited to be on the forefront, pushing the technology to its limits, driving change and delivering solutions that enable new discoveries in research and help advance healthcare to create a better future for all of us.
Click the following links to learn more about Oragene·Dx , ORAcollect, and prepIT·Q2A or send us an email at info@dnagenotek.com.
Click the link to learn more about Twist Bioscience NGS target enrichment solution.
Reference
[1] https://bmcmedgenomics.biomedcentral.com/articles/10.1186/s12920-019-0527-2
[2] https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0138314

